Next‐generation disease modeling with direct conversion: a new path to old neurons - Traxler - 2019 - FEBS Letters - Wiley Online Library
Diversity Arrays Technology (DArT) Marker Platforms for Diversity Analysis and Linkage Mapping in a Complex Crop, the Octoploid Cultivated Strawberry (Fragaria × ananassa) | PLOS ONE
Diversity Arrays Technology (DArT) Marker Platforms for Diversity Analysis and Linkage Mapping in a Complex Crop, the Octoploid Cultivated Strawberry (Fragaria × ananassa) | PLOS ONE

Diversity arrays: a solid state technology for sequence information independent genotyping. | Semantic Scholar

A survey of direct-to-consumer genotype data, and quality control tool (GenomePrep) for research - ScienceDirect

Method of manufacturing a microarray with diversity array technology... | Download Scientific Diagram
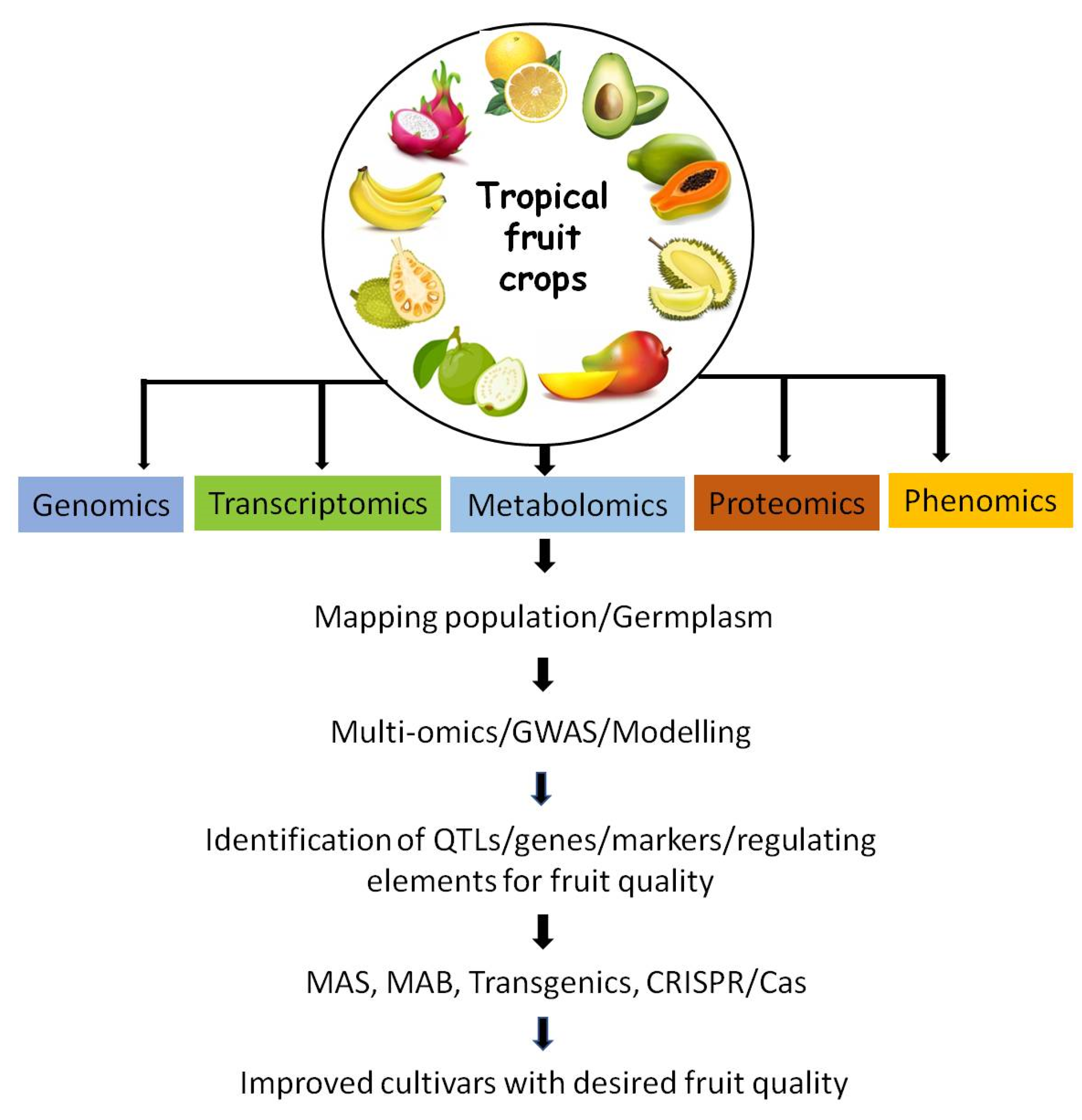
Genes | Free Full-Text | Genomic Approaches for Improvement of Tropical Fruits: Fruit Quality, Shelf Life and Nutrient Content

Figure 10.1 from Diversity Arrays Technology: A Novel Tool for Harnessing the Genetic Potential of Orphan Crops | Semantic Scholar

A 62K genic-SNP chip array for genetic studies and breeding applications in pigeonpea (Cajanus cajan L. Millsp.) | Scientific Reports
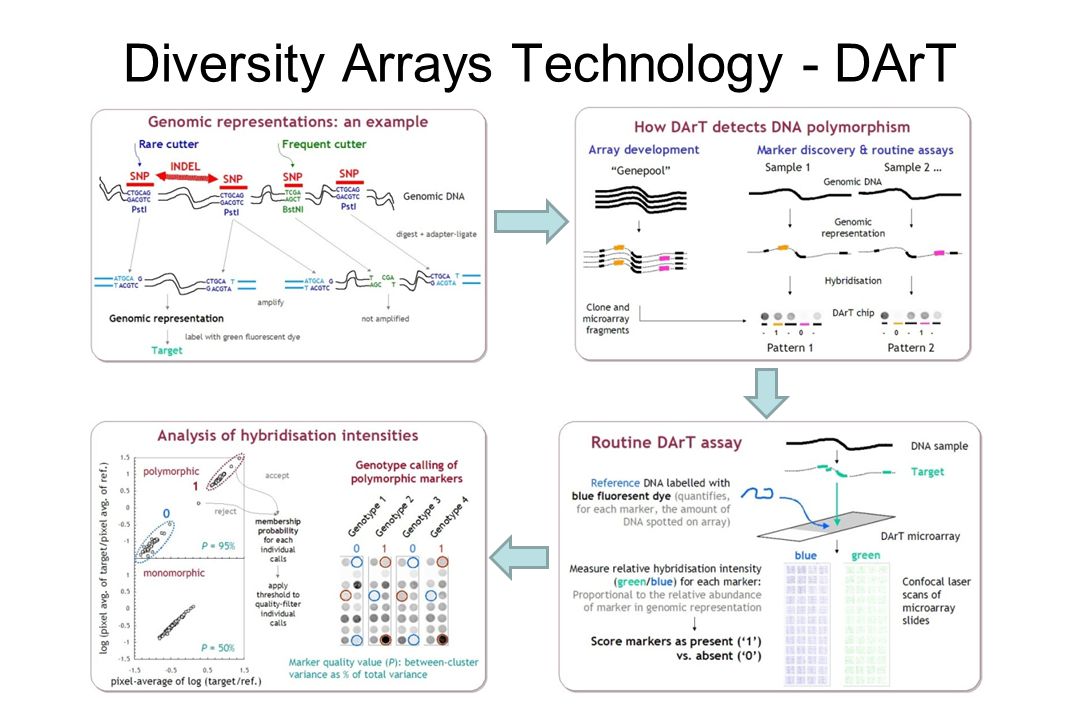
Structural, functional Genome, Transcriptome, Proteome, Metabolome, Interactome Genomics. - ppt download
Diversity Arrays Technology (DArT) Marker Platforms for Diversity Analysis and Linkage Mapping in a Complex Crop, the Octoploid Cultivated Strawberry (Fragaria × ananassa) | PLOS ONE

CRISPR-Mediated Base Conversion Allows Discriminatory Depletion of Endogenous T Cell Receptors for Enhanced Synthetic Immunity: Molecular Therapy - Methods & Clinical Development
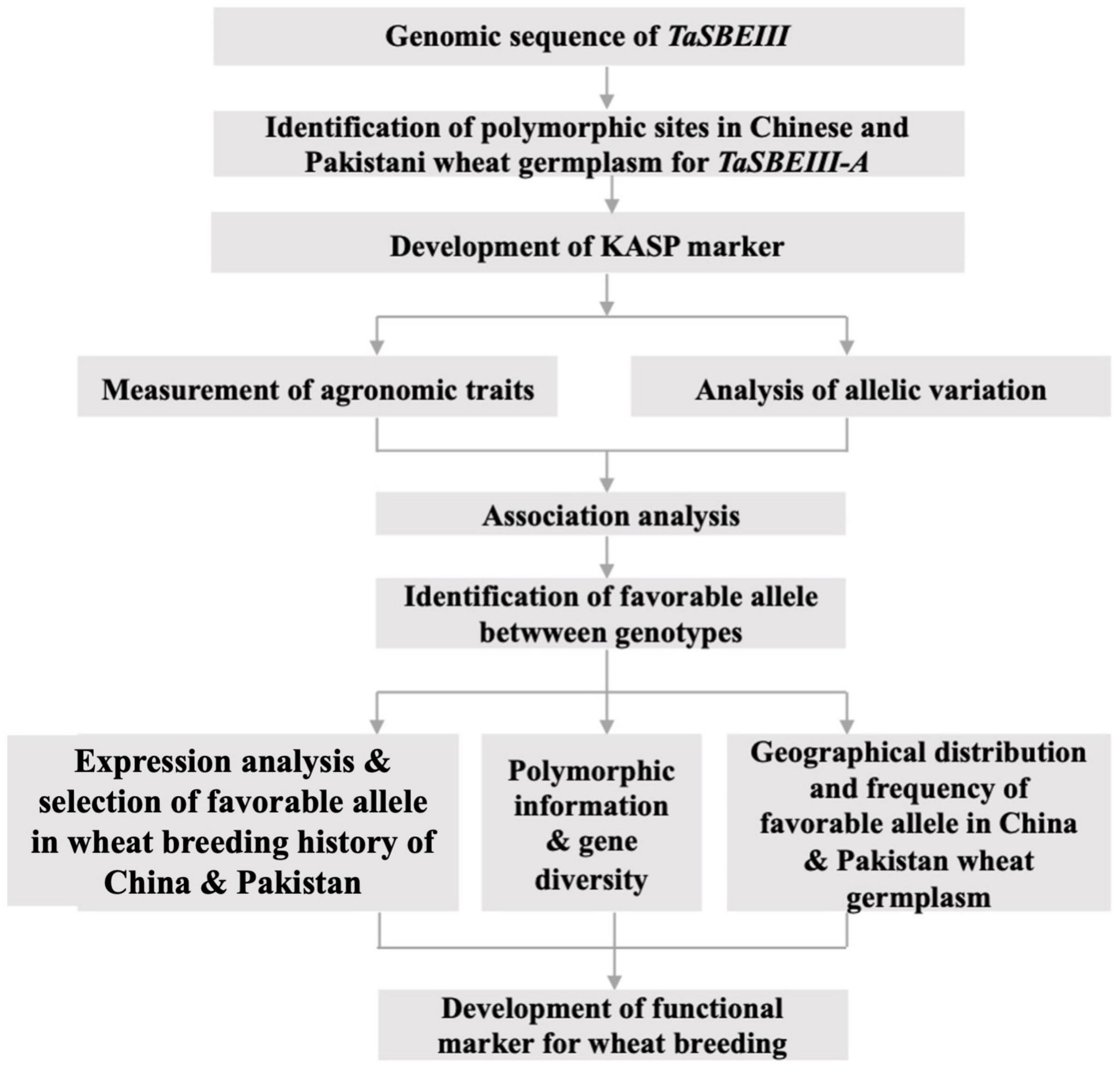
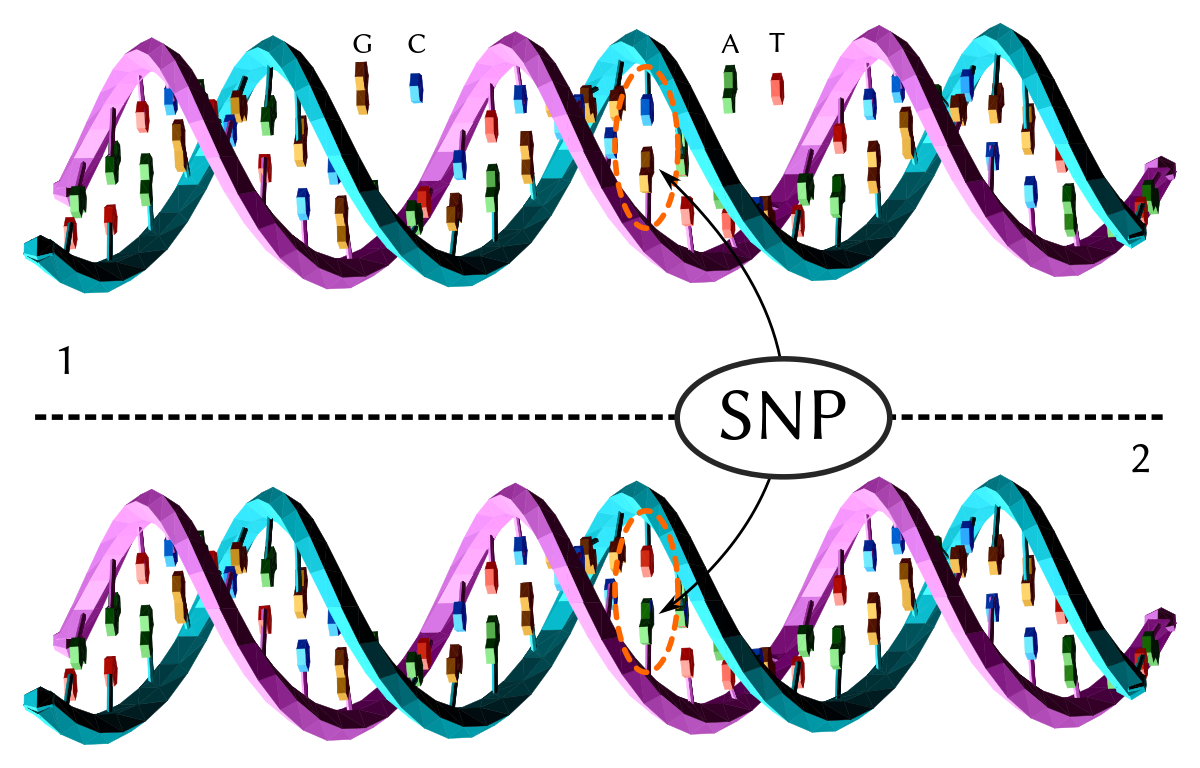
Frontiers | Identification of Single Nucleotide Polymorphism in TaSBEIII and Development of KASP Marker Associated With Grain Weight in Wheat

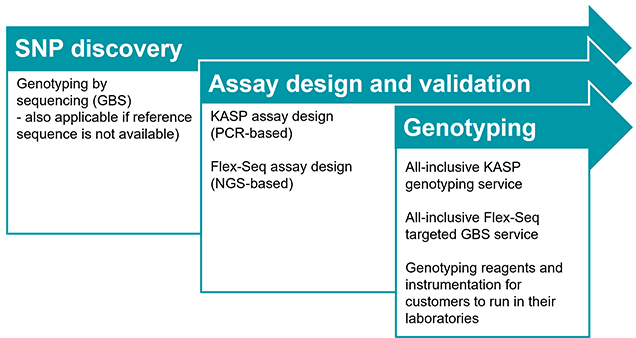
Oat Molecular Markers: Status and Opportunities Howard W. Rines USDA-ARS and Department of Agronomy and Plant Genetics, University of Minnesota, St. Paul, - ppt download

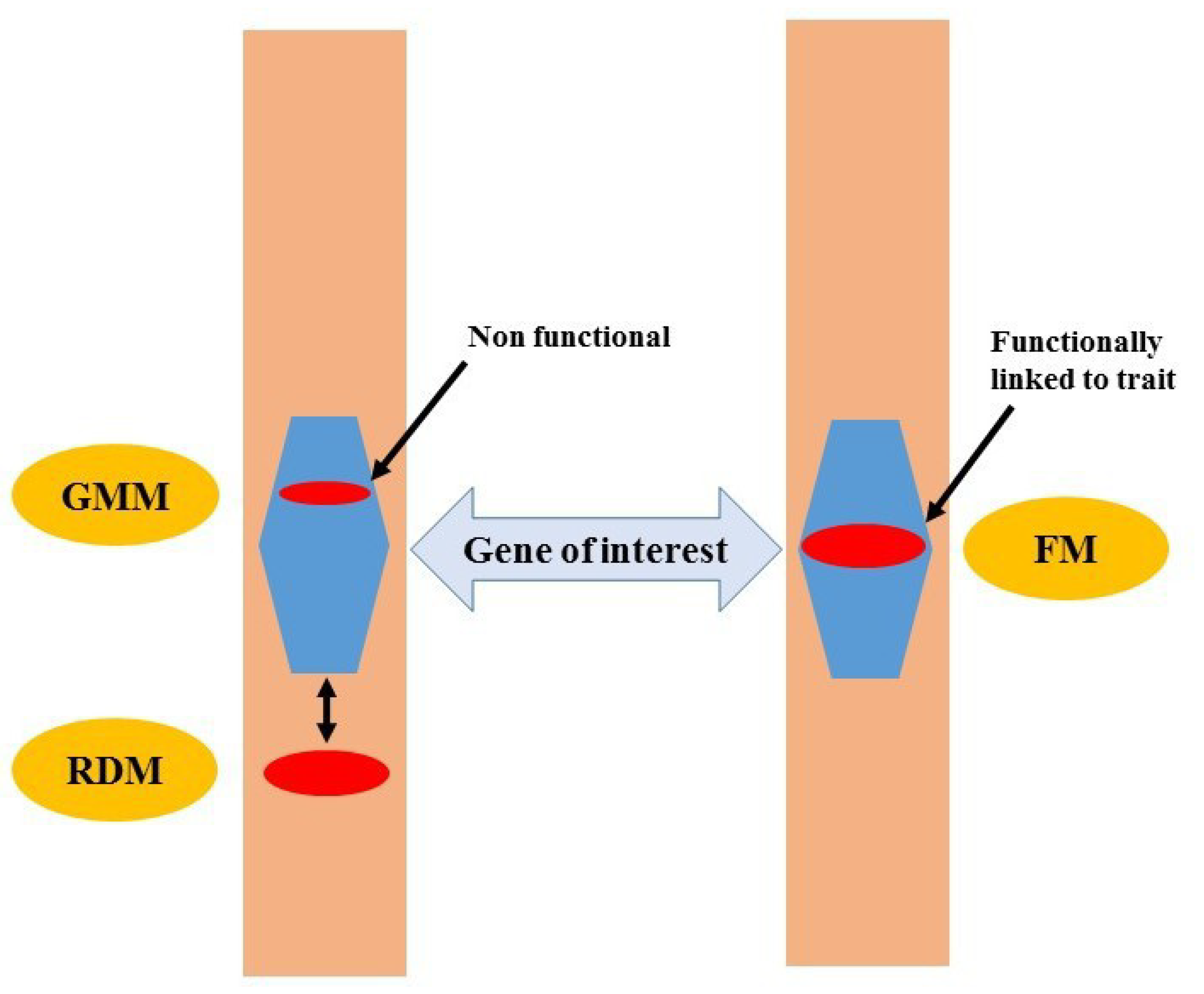
PDF) Conversion of a diversity arrays technology marker differentiating wild and cultivated carrots to a co-dominant cleaved amplified polymorphic site marker | Dariusz Grzebelus - Academia.edu